Tapeworm removed from UK resident’s brain reveals genetic secrets of an elusive Far East parasite
For the first time, the genome of a rarely seen tapeworm has been sequenced. The genetic information of this invasive parasite, which lived for four years in a UK resident’s brain, offers new opportunities to diagnose and treat this invasive parasite.
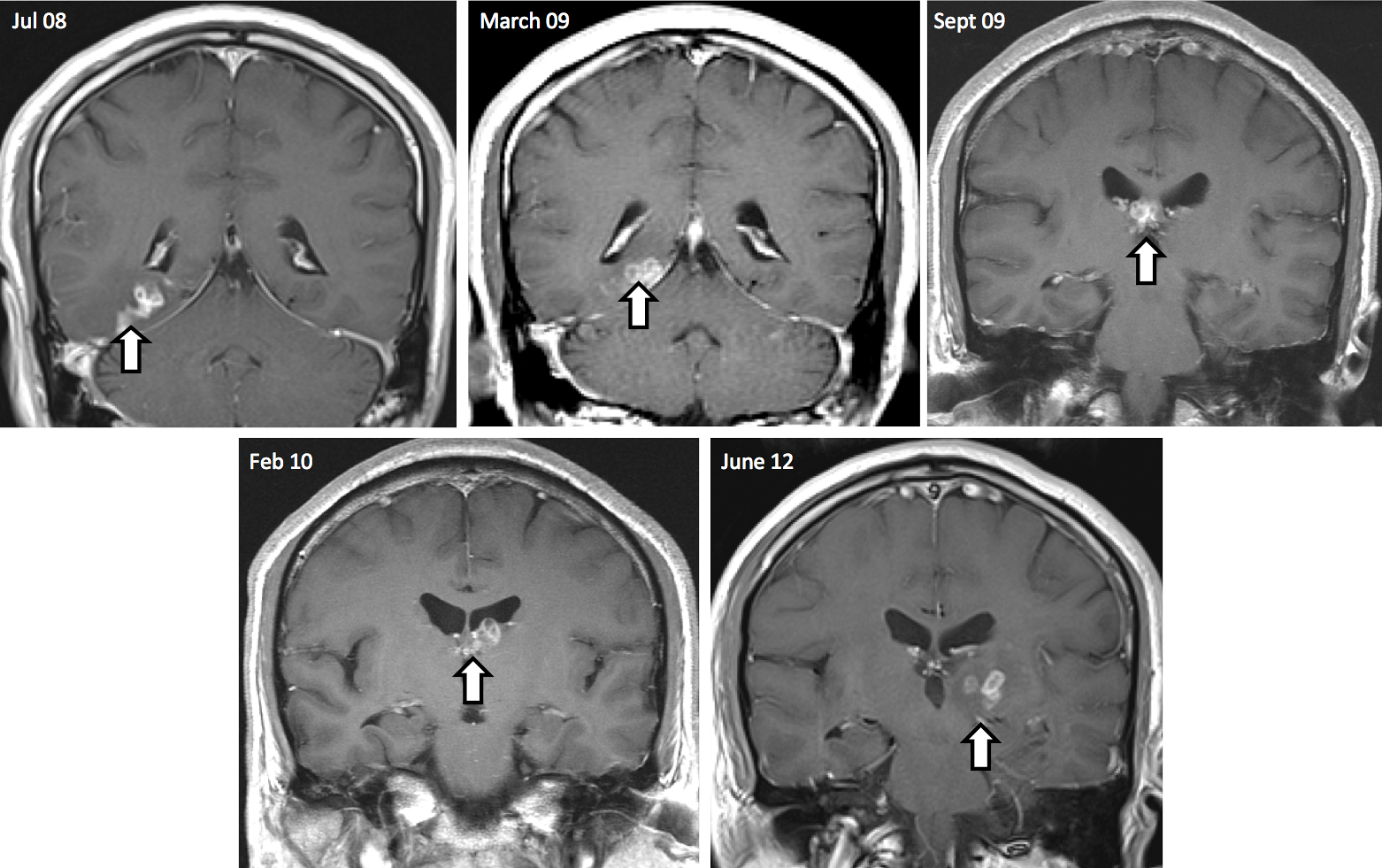
The tapeworm, Spirometra erinaceieuropaei, has been reported only 300 times worldwide since 1953 and has never been seen before in the UK. The worm causes sparganosis: inflammation of the body’s tissues in response to the parasite. When this occurs in the brain, it can cause seizures, memory loss and headaches. The worm’s rarity means that little is known about its complex lifecycle and biology, however it is thought that people may be infected by accidently consuming tiny infected crustaceans from lakes, eating raw meat from reptiles and amphibians, or by using a raw frog poultice – a Chinese remedy to calm sore eyes.
Before the 1cm-long parasite was diagnosed and successfully removed by surgery, it had travelled 5cm from the right side of the brain to the left. The tapeworm was placed on a histology slide by the hospital to confirm the clinical diagnosis. The patient is now systemically well.
“The clinical histology slide offered us a great opportunity to generate the first genome sequence of this elusive class of tapeworms,” says Dr Hayley Bennett, first author of the study from the Wellcome Trust Sanger Institute. “However, we only had a minute amount of DNA available to work with – just 40 billionths of a gram. So we had to make difficult decisions as to what we wanted to find out from the DNA we had.”
To identify the exact species of worm, the researchers sequenced one particular gene, the so-called ‘barcode of life’. Fortunately for the patient, the gene’s DNA sequence revealed that the parasite was the more benign of the two sparganosis-causing worm species. Remarkably, the team also were able to generate sufficient DNA sequence data using standard next-generation sequencing techniques to piece together a draft genome. This is now being used to investigate known and potential treatment targets, which may help patients in the future.
” For this uncharted group of tapeworms, this is the first genome to be sequenced and has allowed us to make some predictions about the likely activity of known drugs. ” Dr Matt Berriman
“We did not expect to see an infection of this kind in the UK, but global travel means that unfamiliar parasites do sometimes appear,” says Dr Effrossyni Gkrania-Klotsas, study author from the Department of Infectious Disease, Addenbrooke’s NHS Trust. “We can now diagnose sparganosis using MRI scans, but this does not give us the information we need to identify the exact tapeworm species and its vulnerabilities. Our work shows that, even with only tiny amounts of DNA from clinical samples, we can find out all we need to identify and characterise the parasite.
“This emphasises just how important a global database of worm genomes is to allow us to identify the parasite and determine the best course of treatment. Additionally, this information can be paired with our work in global travellers’ infection to give additional insights in what infections other patients can get in specific destinations. We are really lucky to be able to work closely with such an excellent facility as the Wellcome Trust Sanger Institute.”
Spirometra erinaceieuropaei’s genome is 1.26Gb long, making it ten times larger than other tapeworm genomes and one-third the size of the human genome. Some of this seems to come from an increase in the number of genes that may help the parasite to break up proteins and invade its host, coupled with the fact that the genome is much more repetitive than other tapeworm genomes.
The team also used this draft sequence to look for similarities and differences from other, previously sequenced, tapeworm species in the GeneDB pathogen database. This has revealed more about Spirometra erinaceieuropaei’s biology than ever before. For example, the worm has a large selection of molecular motors for moving proteins around the cell, which could underpin the large changes in body shape and environmental adaptions that the worm undergoes during its complicated lifecycle.
“For this uncharted group of tapeworms, this is the first genome to be sequenced and has allowed us to make some predictions about the likely activity of known drugs,” says Dr Matt Berriman, senior author and member of Faculty of the Sanger Institute. “The genome sequence suggests that the parasite is naturally resistant to albendazole – an existing anti-tapeworm drug. However, many new drug targets that are being explored for other tapeworms are present in this parasite and could offer future clinical possibilities.”
Because such an elusive worm is rare, discovering targets in the genome to existing licensed drugs could prove to be the best way to treat this rare disease. These data contribute to the growing global database for identifying parasites and parasite provenance and will serve as a resource for identifying new treatments for sparganosis.
Source
[ads1]
Leave a Reply